Key Takeaways
- Consistently high gene detection across multiple donors
- Clustering validates detection of all principle brain cell types
The brain, with its countless interconnected cells and vast networks is a focal point for detailed research. Afflictions like Alzheimer’s disease, schizophrenia, Huntington’s disease, result in some of the most tragic human conditions. Single cell transcriptomic analysis provides fundamental perspectives into the biology of this intricate organ.
Here, four brain tissues were collected from adult C57/BL6 mice by Transnetyx, flash frozen, and shipped on dry ice. Samples were dissociated into single nuclei with Miltenyi Biotec gentleMACSTM dissociator. Mouse brain nuclei were fixed with Evercode Nuclei Fixation v3 kit and frozen at -80C. Whole transcriptome sublibraries were generated with Evercode WT v3.
Whole transcriptome sublibraries were sequenced on an Illumina Novaseq 6000 with a S4 flow cell. Data for the resulting 51,740 nuclei were processed with Parse Biosciences data analysis pipeline v1.2.0.
Consistently sensitive gene detection was achieved as median genes per cell were greater than 2,000 for all samples at a read depth of 32,000 mean reads per cell. Clustering of cells with Seurat v4.0 and additional analysis revealed the expected neuronal and non-neuronal cell types, as shown in the UMAP below.
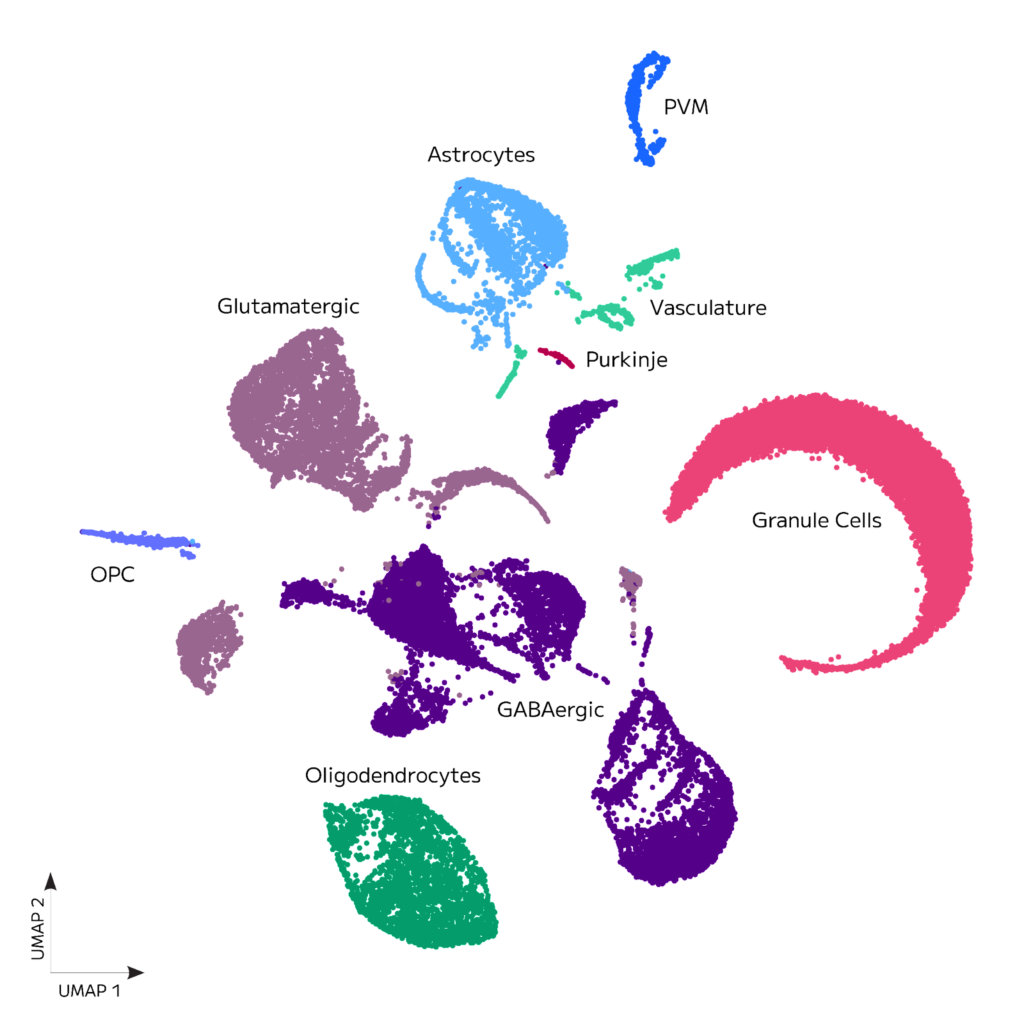
Figure 1. Adult Mouse Brain Nuclei Clustering. 51,740 adult mouse brain nuclei from 4 donors were processed with a single Evercode WT v3 kit.
Next Steps
- Download the data files generated from the preparation and analysis of the adult mouse brain nuclei samples.
We're your partners in single cell
Reach out for a quote or for help planning your next experiment.