Key Takeaways
- High resolution with distinct atrial and ventricular cardiomyocytes cluster separation
- Compatible with whole cells from fresh neonatal mouse heart
- Observation of the expected major cell types with known cell markers
The heart is the first functional organ to develop and the process of heart development is highly conserved between mammalian species. By obtaining the whole transcriptome profiles of single cells in the heart, researchers can gain new insights into the roles of individual cell types in cardiovascular development and disease.
Here, fresh heart tissue was collected from a neonatal (P0) C57/BL6 mouse by Transnetyx and shipped for single cell gene expression experiments. After the enzymatic tissue dissociation, cells were strained, centrifuged, and resuspended in HBSS (Hanks Balanced Salt Solution). The single cell suspension was fixed with Evercode Cell Fixation v2 and whole transcriptome libraries generated with Evercode WT Mini v2.
Whole transcriptome libraries were sequenced on an Illumina Nextseq 550 high-output flow cell, and data for the resulting 9,243 cells were processed with Parse Biosciences Analysis Pipeline v1.0.3. Mean reads per cell were 44,635 while median genes per cell were 3,184. Clustering of cells with Seurat v4.0 and additional analysis revealed the expected cell types and cell marker expression profiles, as shown in the stacked dot plot and UMAP below.
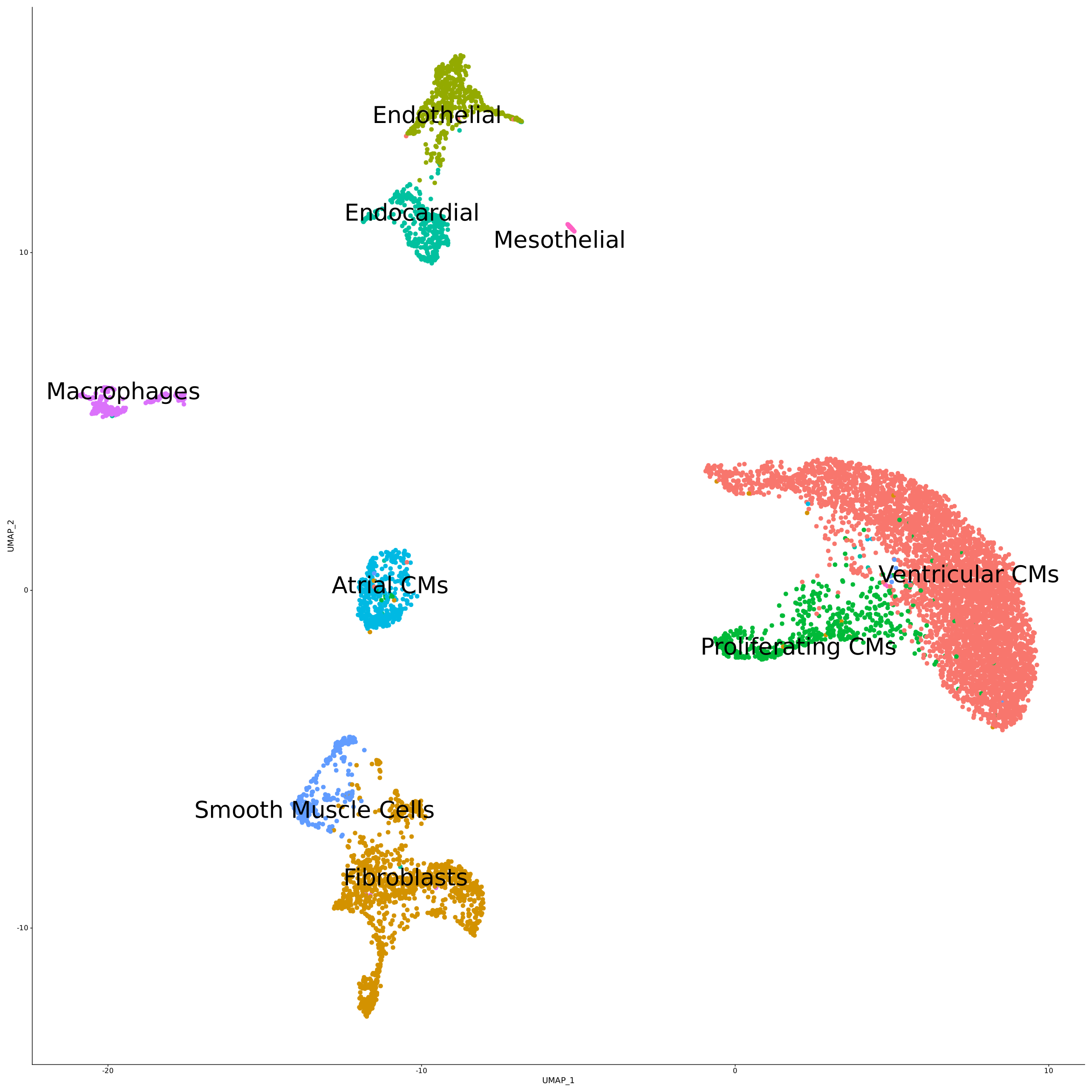
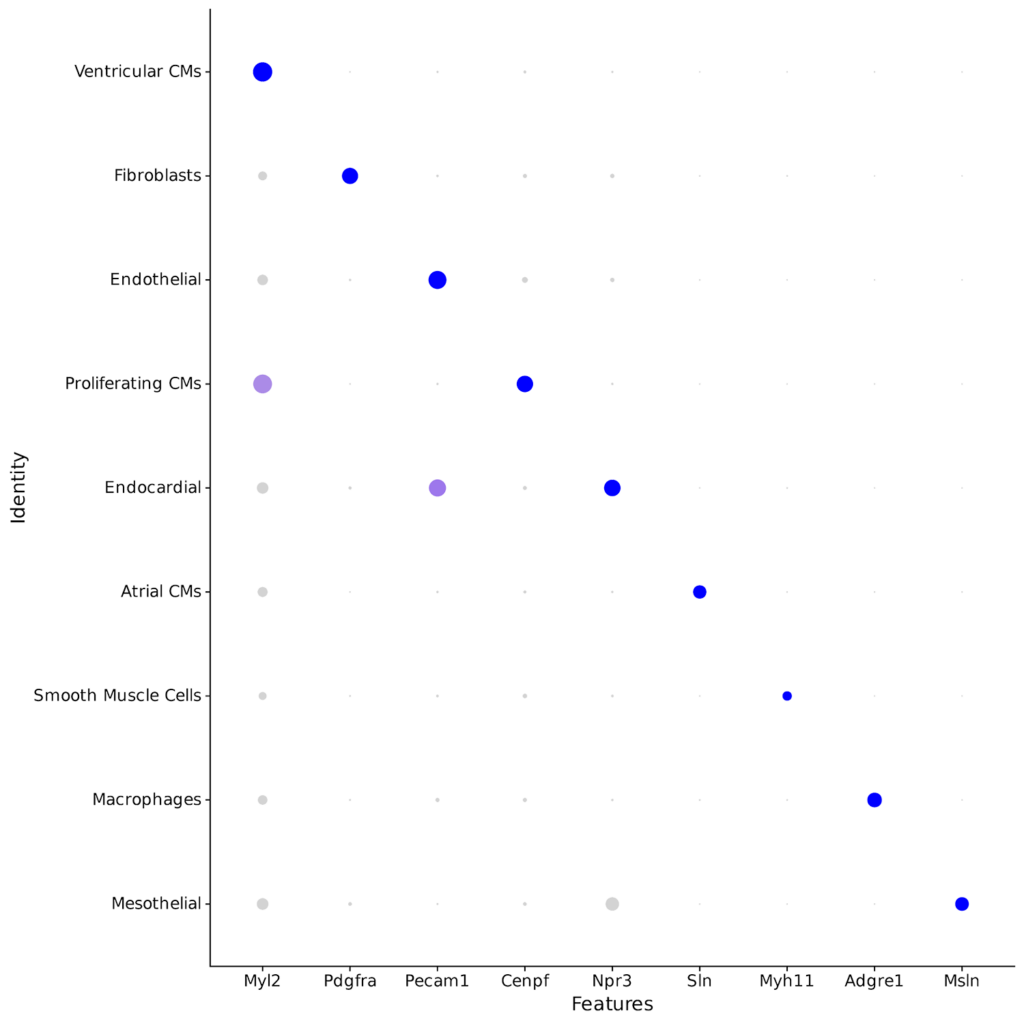
You can further explore the data and do your own analysis by downloading the raw data below.
We're your partners in single cell
Reach out for a quote or for help planning your next experiment.