1 Million Human PBMCs in a Single Whole Transcriptome Experiment
Key Takeaways
- Results from 1 million cells in less than a week
- Analyzing more cells is important to capturing the full complexity of heterogeneous samples
Experimental Design
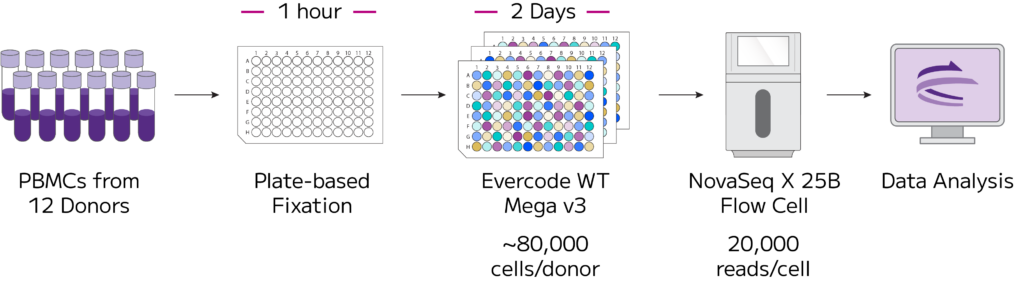
Figure 1: Overview of Experimental Design. Approximately 1 million cells from twelve healthy PBMC donors were processed with the Evercode WT Mega v3 single cell RNA-Seq workflow in a single experiment.
Cryopreserved PBMCs from 12 healthy donors were purchased from a commercial vendor. Samples were thawed in a 37༠C water bath, transferred to a 50 mL centrifuge tube, diluted dropwise with warm FBS media, centrifuged, and washed with cold FBS. All samples had a viability >90% after thawing.
For each donor, approximately 1 million cells were added to 4 wells of a 96 deep well plate. These 48 samples were fixed with the Evercode Cell Fixation v3 High Throughput Plate-Based Workflow, aliquoted into PCR plates, and stored at -80༠C. On a day prior to running the downstream Evercode WT Mega v3 kit, aliquots of fixed sample were thawed in a 37༠C water bath and counted in batches. The median retention across the 48 samples was 46%.
Fixed samples were processed with Evercode WT Mega v3. After barcoding, 59% of cells were retained. Libraries were sequenced with an Illumina Novaseq X on a 25B flow cell.
After demultiplexing, sequencing data were processed with the Parse Analysis Pipeline v1.2.0. A total of 1,084,202 cells were identified at 19,519 mean reads/cell. Data were integrated with Seurat, filtered to remove low quality cells, cell types classified with Azimuth, and annotations finalized manually. After filtering, samples had an average of ~80,000 cells/sample with a total 955,030 cells in the dataset.
Results
Expected PBMC major cell populations were identified in all 12 donors (Figure 2). Rare cell types that are difficult to distinguish when analyzing fewer cells were also identified, including HSPC (hematopoietic stem and progenitor cells), cDC1 (conventional type 1 dendritic cells), Plasmablast, and ILC (innate lymphoid cells).
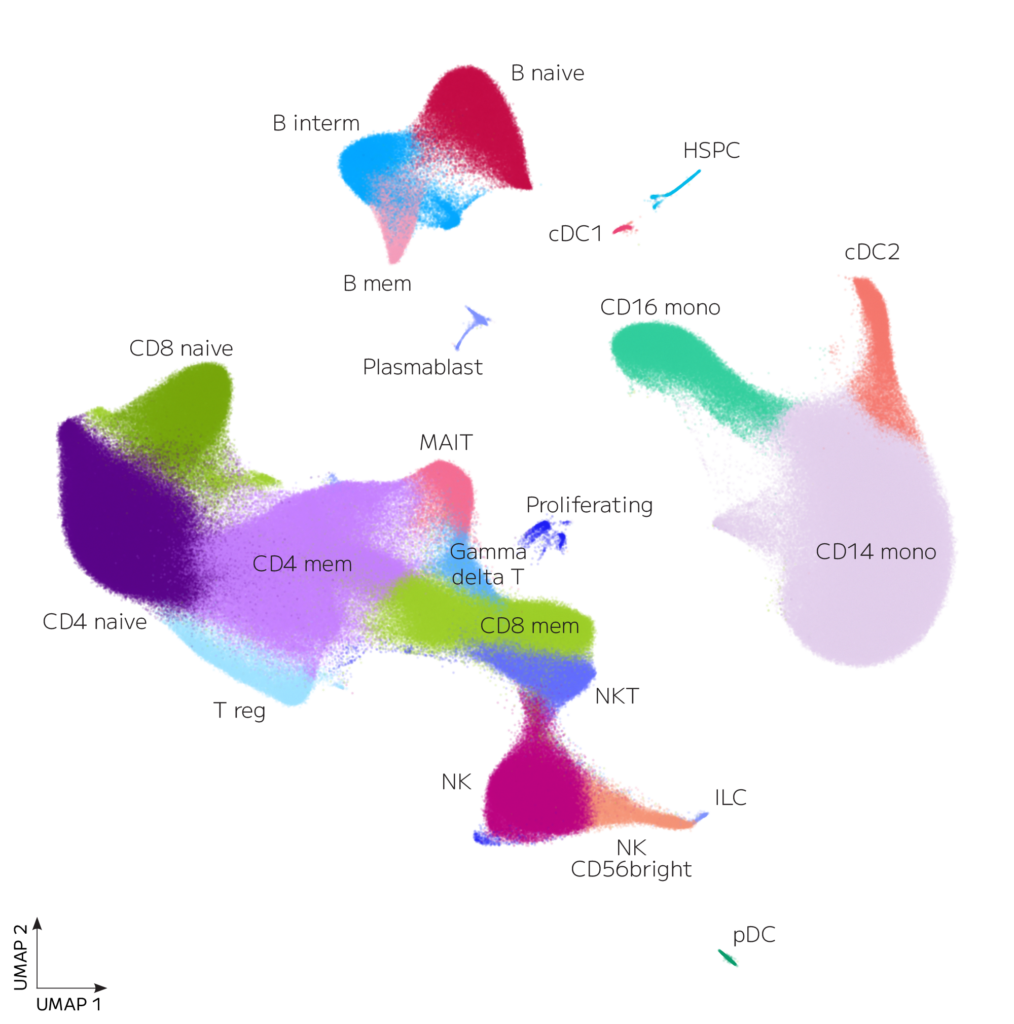
Figure 2: UMAP of 1 million PBMCs from 12 donors. Data from each donor was integrated, clustered with Seurat, and annotated.
The relative abundances of cell types across donors highlights the heterogeneity of PBMC samples, even among healthy donors (Figure 3). This is particularly evident for rare cell types.
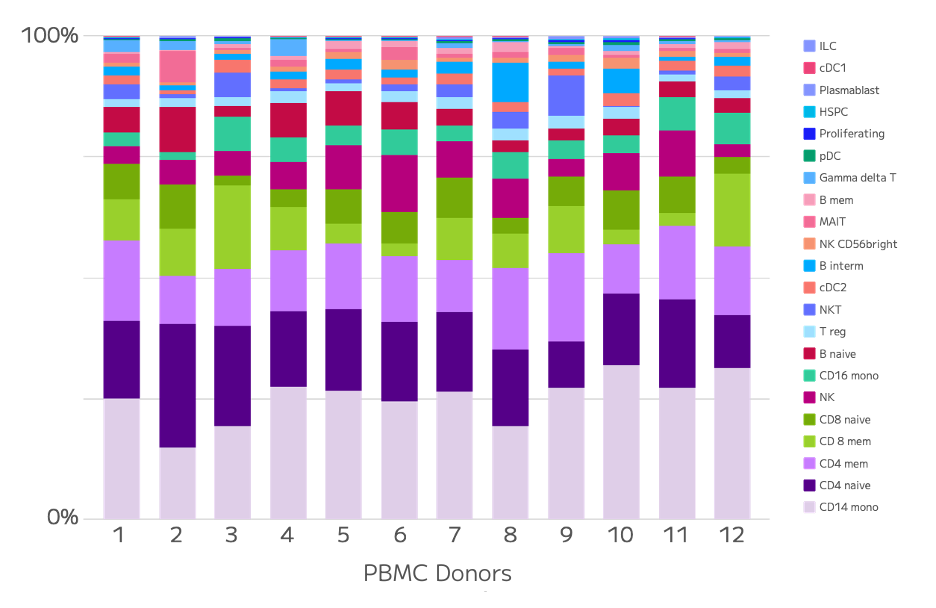
Figure 3: Cell Type Proportions. Relative abundance of each cell type was quantified for each donor.
cDC1s are a subtype of dendritic cells with the lowest abundance with a total of 963 cells in this dataset, accounting for just 0.1% of cells (Table). Dendritic cells are antigen presenting cells that play a key role in the adaptive immune response. Although making up a small fraction of cells in peripheral blood, cDC1s are distinct from other dendritic cells in that they can modulate both CD4+ and CD8+ T cell responses and play an increasingly recognized role in antitumor responses.
Because ~80,000 cells were analyzed for each donor, this dataset identified cDC1 cells in all donors with 38-198 cells per donor. With fewer cells per sample, it would be difficult to resolve this cell type and particularly difficult to make any conclusions about the activity of these cells and dendritic cells as a whole in these samples.
| Cell Type | Total Cells | Cells/Sample | Estimated Cells/Sample with 10,000 Cells |
| Conventional type 1 dendritic cells (cDC1) | 963 | 38-198 | 4-19 |
| Conventional type 2 dendritic cells (cDC2) | 18,077 | 712-2,137 | 82-264 |
| Plasmacytoid dendritic cells (pDC) | 3,306 | 112-454 | 20-63 |
| Total dendritic cells | 22,346 | 1,011-2,672 | 116-335 |
Next Steps
- Download the data files generated from this 1 million cell dataset (below).
This dataset is licensed under the Creative Commons Attribution-NonCommercial 4.0 International (CC BY-NC 4.0) license. Non-commercial users are free to share and adapt the data by providing appropriate credit – Parse Biosciences citation guidelines are available here. If you’re interested in licensing the data for commercial purposes, visit our Dataset Commercial Licensing page.
Downloads
We're your partners in single cell
Reach out for a quote or for help planning your next experiment.