Whole Transcriptome and BCR sequencing: characterizing the B cell repertoire
The Boyd Lab at Stanford studies both the healthy and pathogenic immune response in humans. Here they compare the cellular makeup and BCR repertoire of PBMCs and spleen from a healthy human donor. Samples were fixed and then enriched for B cells using a negative selection kit.
The resulting transcriptome data showed high transcript and gene detection across 87,000 identified cells.
Alongside high-quality whole transcriptome data, they used Evercode BCR to generate paired heavy and light chain sequences across 82,000 B cells, including low abundance B cell types like plasma cells.
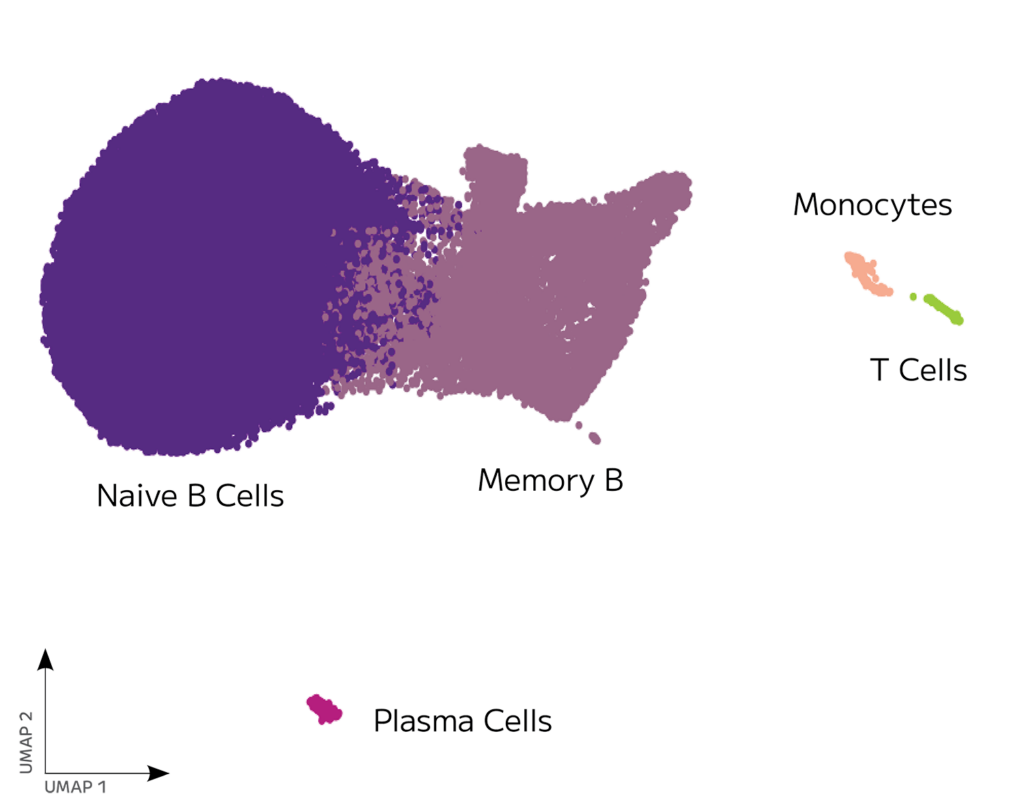
Stimulated B cells can differentiate and undergo isotype switching resulting in both memory B cells and antibody secreting plasma cells. Isotype switching retains the variable region, but switches the constant region isotype resulting in antibodies with different biological responses.
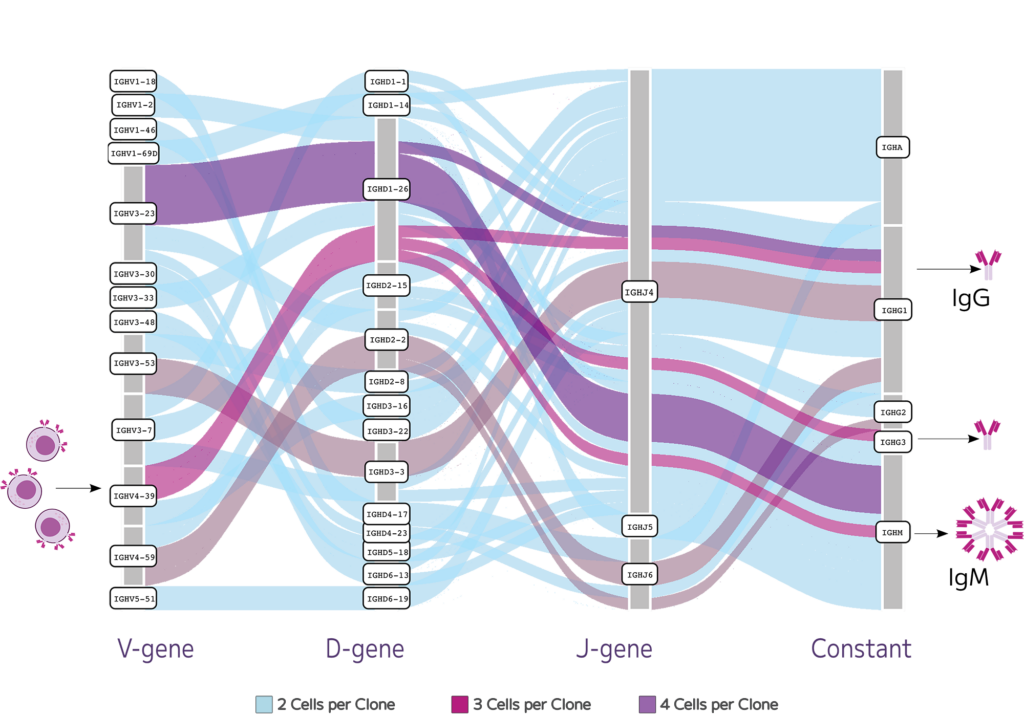
Evercode Fixation preserves fragile cell types like plasma cells, while full-length BCR transcripts enables capture of the full complexity of the B cell repertoire.

"Exceptional, high-quality paired BCR repertoire data at scale."
We're your partners in single cell
Reach out for a quote or for help planning your next experiment.