Transcriptomic Analysis of a Chemotherapy Resistant Cancer Cell State
Cancer lethality arises from the emergence of therapeutic resistance in the face of an increasing array of drugs and novel regimens targeting everything from proliferation, DNA integrity, metabolism, cell contacts, and more. Our group studies an underappreciated endocycling cancer cell state that contributes to therapeutic resistance in numerous cancer settings. Cells in this state uncouple DNA replication from cell division to obtain abnormally high genomic count and cell size (>70x volume).
We work on a human prostate cancer cell line, chosen because the cells readily become refractory to treatment. Utilizing the Evercode WT v2 assay, we profiled untreated cancer cells alongside those in the cancer cell state. Our first dataset successfully captured the dramatically increased RNA content and provided insight into multiple cell state trajectories after chemotherapy induction.
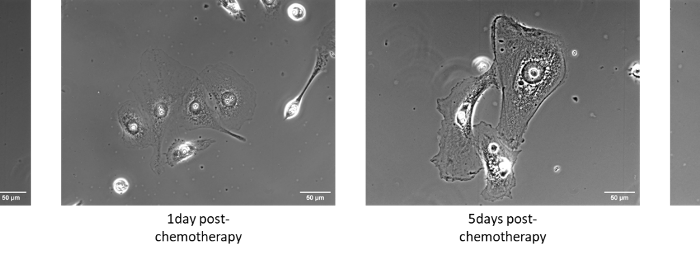
Phase contrast images of samples showing increasing cell size.
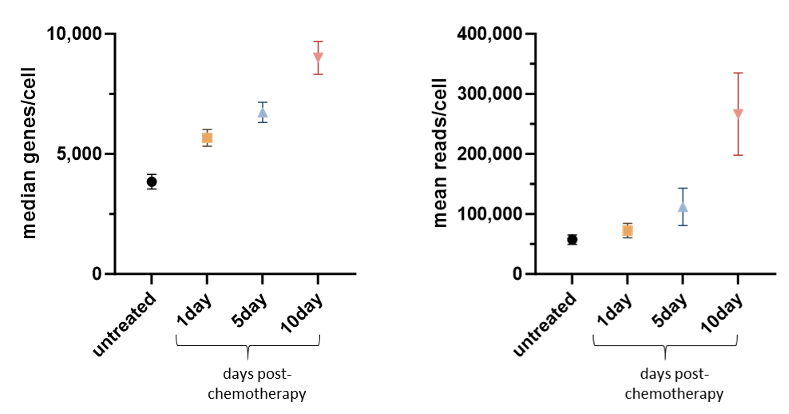
Reads and median genes detected per cell across four sample timepoints. Error bars are standard deviation across three biological replicate samples. Because all cells are represented in each sublibrary, reads/genes/UMI per cell are indicative of RNA amount per cell.
Sample Table
| ID | Sample | Recovery Time post-chemo (days) | Processed |
| A | Untreated Cell | – | Cells |
| B | Treated Cell | 1 | Cells |
| C | Treated Cell | 5 | Cells |
| D | Treated Cell | 10 | Cells |

"Single cell resolution is vital to capturing the complexity of cancer but droplet-based methods are unreliable for morphologically large cells."
We're your partners in single cell
Reach out for a quote or for help planning your next experiment.