Scientists in major universities are already using the Evercode BCR Evercode TCR assays.
The Boyd Lab at Stanford University used the Evercode BCR assay to capture both the cellular makeup and BCR repertoire in healthy people. They fixed the B cells and later ran the assay. The whole transcriptome data showed high transcripts and gene detection across 87,000 cells. The paired heavy and light chains generated across 82,000 B cells featured stimulated B cells that differentiated and underwent isotype switching, resulting in both memory B cells and antibody secreting plasma cells (Fig. 4).
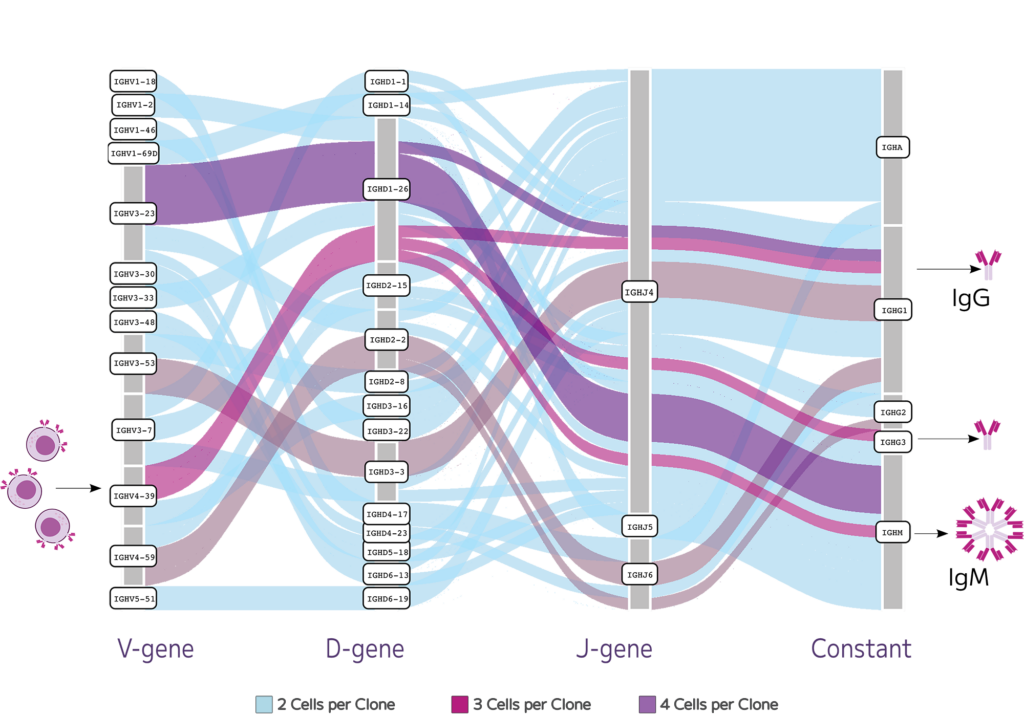
Fig.4: Stimulated B cells can differentiate and undergo isotype switching resulting in both memory B cells and antibody secreting plasma cells. Evercode Fixation preserves fragile cell types like plasma cells, enabling deep profiling of the clonal diversity and providing insight into patient specific immune responses.
The Reticker-Flynn Lab at Stanford is utilizing the Evercode TCR technology to examine the impact of the lymph nodes in tumor metastasis and subsequent changes to the phenotype and clonotype diversity within mouse T cells. The study is ongoing, and the team plans to leverage TCR sequencing data to gain insights into tumor dynamics and activity.
Moreover, in a recent study presented at AAI 2024, Parse scientists demonstrated the Evercode power of scalability by examining BCR clonal diversity in autoimmune diseases using the Evercode BCR assay. They simultaneously profiled both the BCR and whole transcriptome of up to 1 million cells from 24 samples in a single experiment. This approach enabled the detection of all major B cell subtypes in the peripheral blood of 12 subjects with autoimmune diseases and 12 healthy controls. The data illustrated BCR isotypes, captured a majority of cells with a paired chain, detected full length chains, and differentiated dominant and rare clones across a host of autoimmune diseases (Fig. 5).
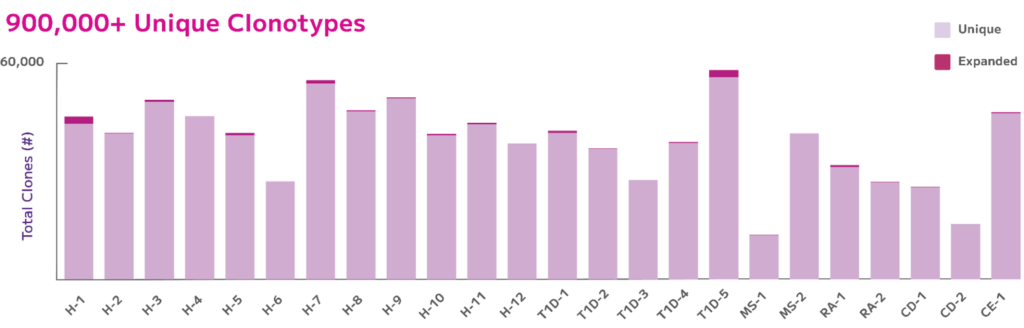
Fig. 5: Number of Unique vs. Expanded Clones. Total unique clones varied between 13,000 – 59,000 per donor where all donors had over 95% unique clones in their clone pool with donor H-1 and T1D-5 having the most expanded clones ~5%.
When focusing on the 5 samples from Type 1 Diabetes donors, the team recognized different immune responses in these patients based on their clonal expansion and isotype switch.
At the same conference, Parse scientists demonstrated the sensitivity, accuracy, and scalability of the Evercode TCR analyzing the effects of aging on mouse T cell receptor diversity using the Evercode TCR assay. By profiling hundreds of thousands of mouse spleen-resident T cells at different life stages, they identified all major T cell populations and their paired TCR sequences. These findings revealed differences in clonotype diversity correlated with age and cell type, highlighting the utility of this technology in studying the impact of aging on the immune system (Fig. 6).
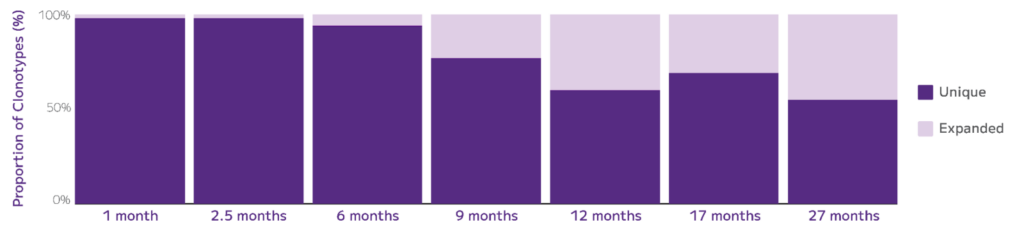
Fig. 6: Evercode TCR for Mouse: proportion of unique to expanded clonotypes over the mouse lifespan. The unique clonotypes (dark purple) are defined as only being detected in 1 cell: their percentages decrease from 98% to 55% over 26 months, demonstrating the T cell repertoire contraction over time.