Performance of Evercode™ WT in Human Kidney Dissociated Tumor Cells (DTCs)
Key takeaways
- Distinct clustering of complex dissociated tumor cells (DTCs)
- 15 RCC samples across four groups processed in single experiment
- Revealing insights into the correlations between four distinct cohorts and cell types
Renal cell carcinomas (RCCs) is the thirteenth most prevalent cancer type worldwide, accounting for 2.4% of all cancers1. RCCs are categorized in three main, distinct subtypes of kidney cancer – each associated with varying pathology and patient outcomes. Clear cell RCC (ccRCC) is the most prevalent subtype, accounting for 75% of cases, followed by papillary (pRCC) and chromophobe (chRCC) subtypes.
To conduct single-cell transcriptomic analysis on the 3 primary RCC subtypes and normal adjacent kidney, we acquired 15 dissociated tumor cell (DTC) samples listed in the table below through BioIVT. DTCs were fixed with Evercode Cell Fixation v2 and whole transcriptome libraries were generated with Evercode WT v2.
Sample Table
| Cohort Type | Stage | Gender | Sample Alias |
|---|---|---|---|
| Clear cell | I | F | RC-2 |
| Clear cell | I | M | RC-3 |
| Clear cell | I | F | RC-11 |
| Clear cell | I | M | RC-16 |
| Clear cell | IA | F | RC-6 |
| Clear cell | III | M | RC-9 |
| Clear cell | III | M | RC-13 |
| Clear cell | III | M | RC-14 |
| Clear cell | III | F | RC-15 |
| Clear cell | NA | F | RC-4 |
| Chromophobe | III | M | RC-12 |
| Normal adjacent | I | M | RN-1 |
| Normal adjacent | I | M | RN-10 |
| Normal adjacent | I | M | RN-17 |
| Papillary | II | F | RC-7 |
| Papillary | II | M | RC-8 |
Whole transcriptome libraries were sequenced on an Illumina Nextseq 550 high-output flow cell, and data for the resulting 31,593 cells were processed with Parse Biosciences data analysis pipeline v1.0.5. Median genes per cell was 1,428 at a read depth of 26,867 mean reads per cell. Clustering of cells with Seurat v4.0 and additional analysis revealed the three RCC subtypes.
Since the main differentiating factor for subtyping of RCC is the cell of origin and associated histology, we investigated the cell type composition of each sample. Interestingly, there is a distinct separation of the chromophobe sample’s cluster in the UMAPs shown below. This cluster exhibited a strong resemblance to cells annotated as collecting duct cells, which holds significant relevance due to the known association of this RCC subtype with its origin from this specific cell type2.
A)
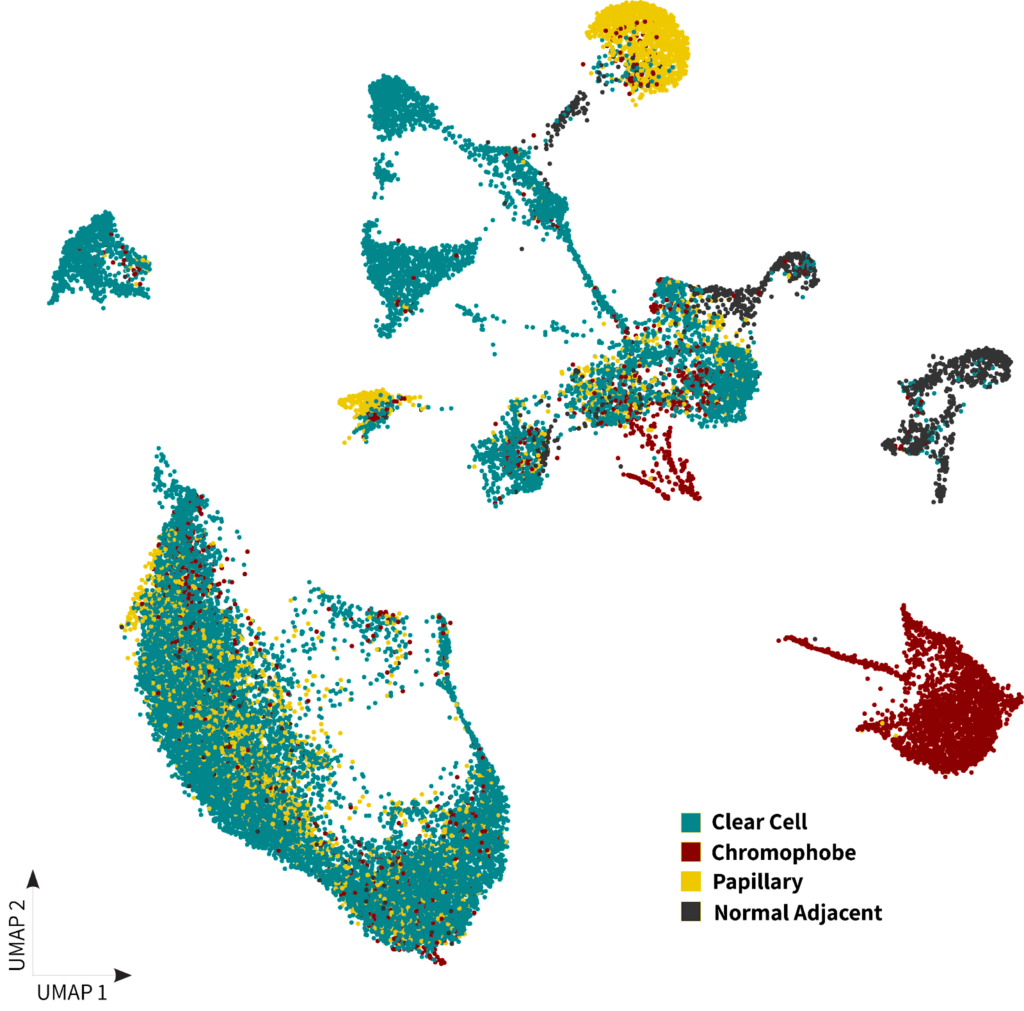
B)
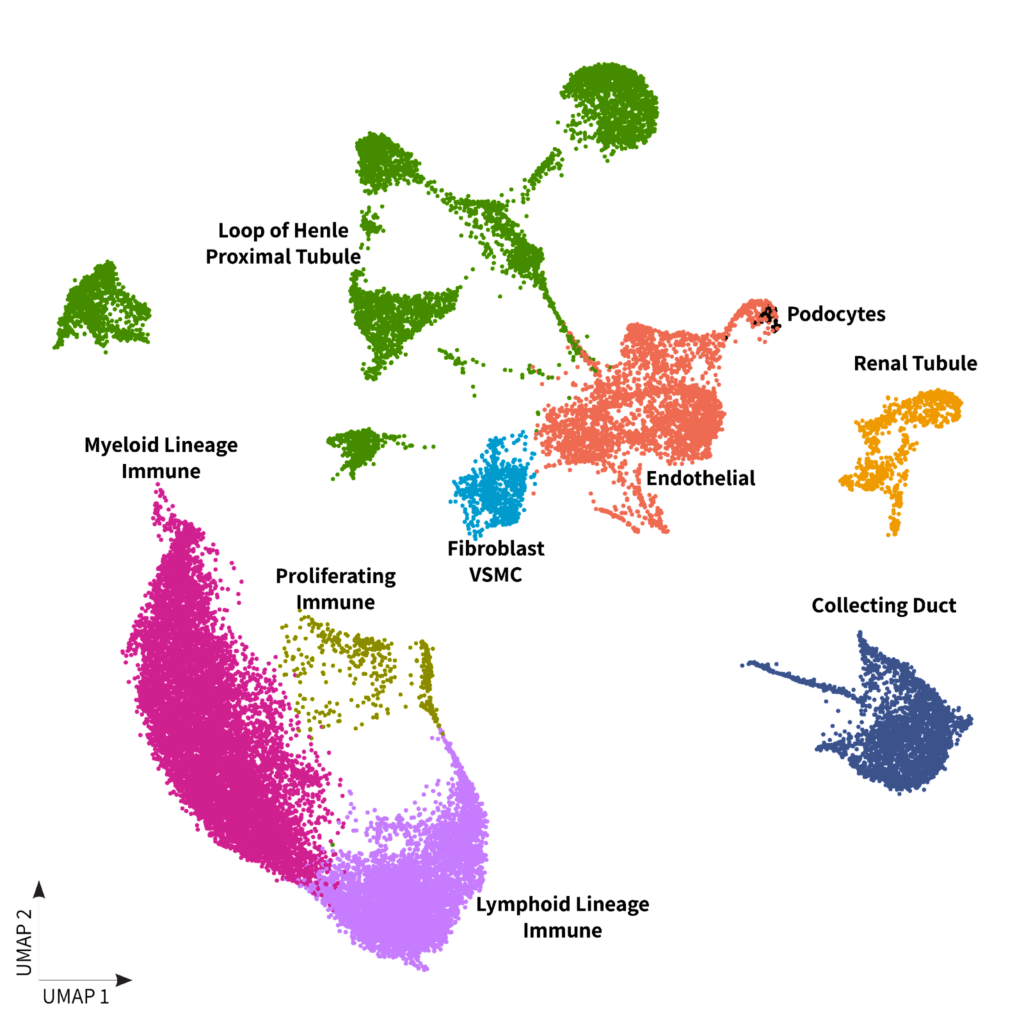
Figure 1: A) Kidney DTCs dataset clustering with cells colored by sample type B) Kidney DTCs dataset clustering with cells colored by cell populations
Moving onto the clear cell and papillary subtypes, our analysis revealed a significant enrichment of proximal tubules and loop of Henle cells. The relationship of these cells with the papillary subtype have been previously reported in the literature2 (2). In addition, both clear cell and papillary subtypes prominently featured immune cells, in contrast to chromophobe, a subtype renowned for its better prognosis.
Finally, we observed that normal adjacent DTC samples formed distinct clusters separate from the cancer samples. These clusters were primarily composed of renal tubule cells as shown in figure 1.
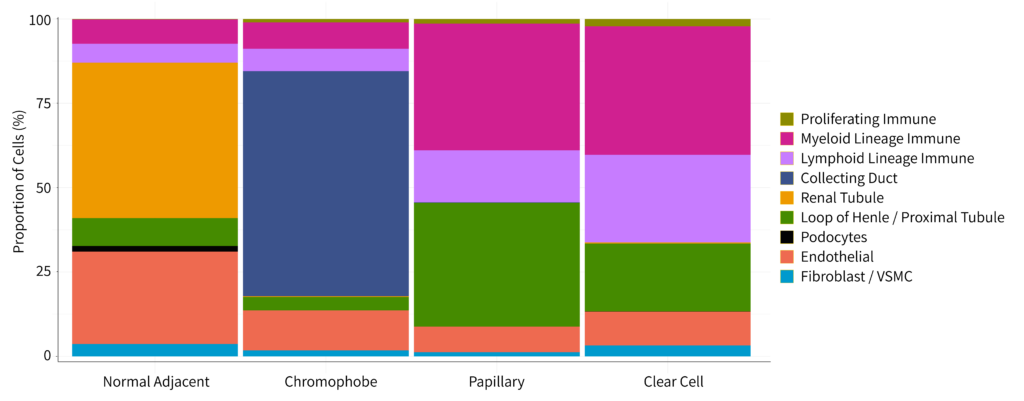
Figure 2: Bar chart with the proportion of each cell type within the four biological groups.
We encourage you to further explore the data and do your own analysis by downloading the raw data below.
References:
We're your partners in single cell
Reach out for a quote or for help planning your next experiment.